Crop Disease 101
Crop Disease: What is it?
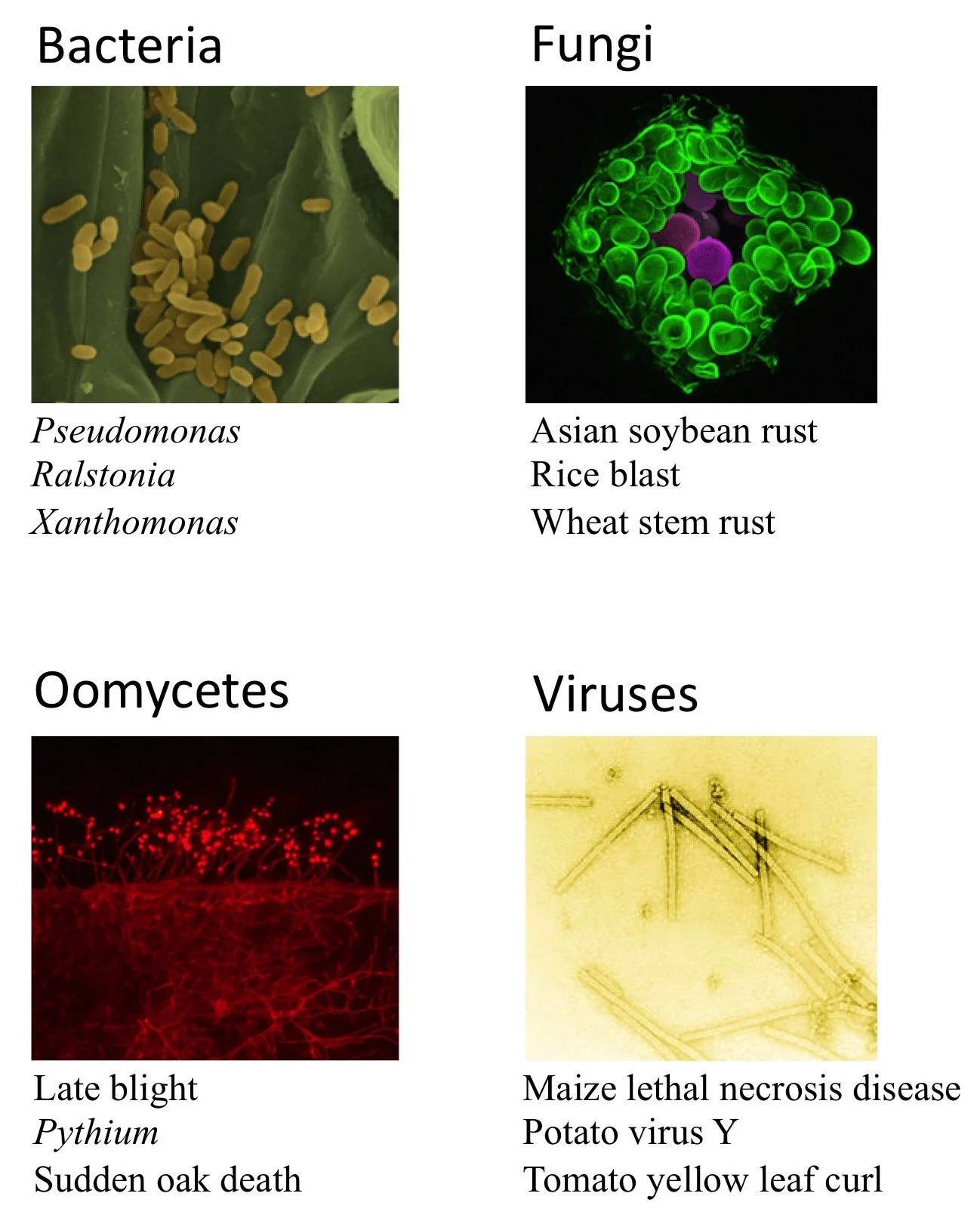
Just like people, plants can get sick too. Plant diseases are caused by four main types of pathogens: bacteria, fungi, oomycetes, and viruses. Of these, fungi are the most prevalent, but all pathogens can be highly lethal to susceptible plants, with losses that can approach 100%.
Plant Immunity 101
by Prof. Jonathan Jones, The Sainsbury Laboratory
Both plants and animal tissues are a rich food source for those microbes that are adapted to exploit them. Colonization by such microbes, and the associated tissue reactions, results in disease. Crop losses to disease cost at least $10B per year and reduce food availability.
Immunity in animals involves production of antibodies, as well as an innate (inherited) immune system that does not depend on immunization, and activates cell defenses upon recognition of molecules shared by many pathogens. More primitive animals, such as insects, only carry an innate immune system and do not have the capacity to make antibodies.
Like insects, plants also rely solely on innate immunity. Plant defense mechanisms are very effective, provided they are activated early enough during the interaction with a disease-causing microbe (a pathogen). Prompt activation depends upon detection of molecules from the pathogen. This detection requires receptor proteins. Some Pattern Recognition Receptors (PRRs) are localized on the surface of cells and detect Pathogen-Associated Molecular Pattern (PAMP) molecules in the spaces between cells. Others reside inside cells and detect pathogen effector molecules injected into plant cells. These intracellular receptors are usually NB-LRR proteins (or simply NLR), so-called because they have a common structure with a Nucleotide-Binding site and Leucine-Rich Repeats. Most plants have hundreds of different NLR-encoding genes. Plants show enormous diversity in their repertoires of NLR genes. Most disease resistance (R) genes used by plant breeders to enhance crop performance usually encode NLRs.
Photo credits: Bacteria – Xanthomonas oryzae on the leaf surface. From Moscou, MJ and Bogdanove, AJ (2009). Science DOI: 10.1126/science.1178817. Reprinted with permission from AAAS; Fungi – Phakospora pachyrhizi breaking through the leaf surface to release spores (purple). Tolga Bozkurt, The Sainsbury Laboratory, Norwich; Oomycetes – Phytophthora infestans colonising a leaf and sporulating at the surface. Sebastian Schornack, The Sainsbury Laboratory, Cambridge; Viruses – Tobacco Mosaic Virus (TMV) particles, Wikimedia, author unknown.
Genes from one plant can help another:
Nearly all plants can benefit from supplementing their capacity to recognize pathogens by the addition of PRRs or NLRs from another plant. 2Blades, and its colleagues in The Sainsbury Lab, Norwich (TSL) and elsewhere, have pioneered such work. For example, pepper carries an NLR gene (Bs2) that confers resistance to bacterial spot caused by Xanthomonas. Tomato is susceptible to bacterial spot, but when the Bs2 gene from pepper is introduced, it is much more resistant, resulting in a doubling of tomato yield in field trials in Florida. The PRR gene from Arabidopsis called EFR enables defense to be activated in response to a PAMP that is not perceived in tomato. When EFR is introduced into tomato, the resulting plants can recognize this additional PAMP, and have elevated resistance to bacterial disease.
Methods of breeding plants with improved disease resistance
by Prof. Brian Staskawicz, University of California, Berkeley
Conventional plant breeding:
Classical plant breeding got its start in the dawn of agriculture over 10,000 years ago, but it only became a formal discipline in the early 1900’s with the first efforts to breed resistance against wheat stripe rust. Sir Rowland Biffen was the first to apply Mendel’s laws to plant breeding, focusing on rust diseases of cereals. Thus, plant breeding as a science has only been in existence for a little over 110 years.
Conventional breeding is carried out by collecting sexually compatible species, intercrossing by transferring pollen from one plant to the other, and selecting desired traits. Some traits, including many disease resistances, are dominant and are seen in the first generation. Conventional plant breeding has been the engine for improving crops since Biffen, but it has limitations. Crops used in crossing must be sexually compatible. Deleterious traits may be linked to the desired trait gene, requiring recurrent selection, also known as back crossing, to maintain the desirable parent type. In some cases, it may be very difficult to break this “linkage drag”. Breeding in some crops may have only a narrow genetic base, and expanding the gene pool into relatives or wild species may offer a greater spectrum of traits. For example, wild species can be a rich source of resistance genes, but sexual compatability with agronomic varieties may be limited to close relatives.
Molecular plant breeding:
Over time, scientists developed the capability to identify markers near genes. In the early 1980s, the study of molecular biology improved the understanding of genes, and it was possible to link observed traits with molecular markers. New and better methods were quickly developed for many crops.
Tools to sequence entire genomes, all of the genes in a particular organism, developed next, and it became possible to find small variations in DNA that can be used as gene markers. The advantage of this approach is that one can take a little tissue from a seed or small seedling and rapidly identify the specific plants with the desired genes without needing to grow the plant to full size.
The next development was the improvement of the means to transfer genes between plants that are not interfertile. It was discovered that a soil bacterium, Agrobacterium tumefaciens, can transfer genes into plants, and scientists use this method to expand the ways genes may be crossed into plants.
Scientists have explored natural variation and found that some genes in plants may be used by the pathogen in its life cycle, causing susceptibility. The tools of gene editing now enable the tailored alteration of sequences within genes, for example to limit the vulnerabilities caused by susceptibility genes.
Currently, scientists are gaining the ability in plant breeding to introduce multiple individual genes in defined regions, together, to act as a single locus, so that they don’t segregate and break down. This way a cassette of genes can be transferred to other varieties easily through breeding and the resistance may be durable.
Glossary
Effectors: Proteins produced by pathogens that are introduced directly into plant cells to manipulate the plant or overcome plant defenses.
Gene editing: A method for making precise small changes in DNA through the introduction of enzymes attached to DNA-binding proteins that cut DNA at specific sequences.
Nucleotide-Binding Leucine Rich Repeat Receptor (NB-LRR or NLR): The most common class of plant immune receptor. These receptors are intracellular proteins that recognize virulence factors (effectors) produced by pathogens, or the indirect effects of their action such as changes to host proteins.
PAMP: An acronym that stands for Pathogen-Associated Molecular Pattern. This refers to any molecule produced by a pathogen that can be recognized by a plant as “foreign.” For instance, fungal cell walls are made of chitin, which is not present in plants, and chitin is recognized as a PAMP. Other known PAMPs are proteins or protein fragments such as those from bacterial flagella.
Pathogen: a disease-causing organism. Many fungi, bacteria, oomycetes, and viruses are pathogens of plants.
Pattern Recognition Receptor (PRR): a receptor that recognizes a molecule produced by a pathogen (a PAMP). These receptors are located on the surface of plant cells.
Resistance (R) gene: a gene encoding a protein that produces resistance to a particular pathogen. Most R genes are NLR or PRR receptors, but there are exceptions.
Transcriptional Activator-Like Effectors (TALEs): A special class of effectors produced by some Xanthomonas pathogens that can bind DNA and activate host genes. The TAL proteins have a repeat structure that encodes the DNA binding specificity through two specific amino acids in each repeat. Because this code is understood, artificial TALEs can be engineered to bind any given DNA sequence, and TALE nucleases (TALENs) are used in gene editing.