Related news
April 5, 2024
2Blades Receives Congressional Funding to Advance Corn Mycotoxin ResearchFebruary 14, 2024
2Blades Delivers on Project with Bayer Crop Science In Effort to Combat Asian Soybean Rust
Nucleotide-binding Leucine-rich Repeat (NLR) proteins are plant receptors that recognize “effector” proteins introduced inside the plant cell by a pathogen. Pathogens use these effectors to suppress plant defenses, but plants that have the right NLR can detect specific effectors and switch on a powerful immune response. Plants with multiple NLR receptors against a pathogen can have strong, long-lasting resistance to disease – protecting crops and reducing the need for crop chemicals.
Yet individual plants may not have NLRs to a specific pathogen and will succumb to disease. Since the earliest days of agriculture, farmers have found they can select resistant plants. Through plant science we have come to understand that in this way NLRs were bred into crops, and today, with modern breeding tools, individual specific NLRs can be precisely selected and introduced into plants. Still, conventional approaches for identifying functional NLRs are labor intensive, expensive, and require advanced expertise.
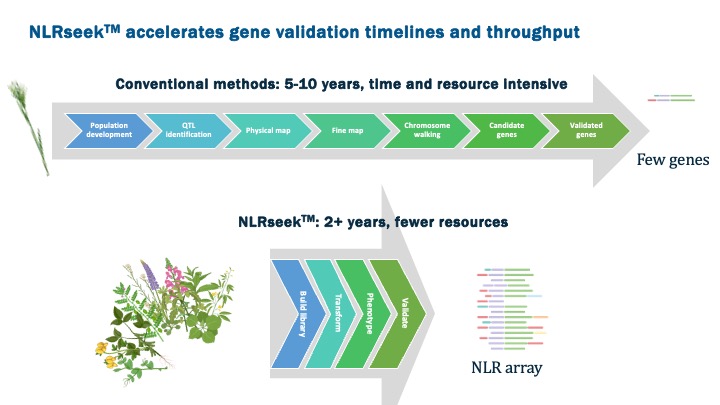
The NLRseekTM program was established to explore observations made by Dr. Matt Moscou about NLR genes that suggested that certain highly expressed genes provided a means to identify high-performing NLRs more readily.
To test this idea, 2Blades undertook a process of selecting 1000 promising NLR genes from a diverse panel of grasses and worked with a team of transformation scientists at Kaneka Corporation to introduce them one at a time into wheat plants, producing a living array of wheat plants differing by a single NLR. This array was then screened with a series of wheat pathogens and found to vastly accelerate the identification and validation of useful disease resistance genes against several of the most damaging diseases of wheat, a critical global food crop.
The insight into the unique expression pattern of high functioning NLR genes contradicted the general body of knowledge about how these genes work. By implementing this expression signature, together with the high throughput wheat transformation capability of Kaneka, we assembled a wheat array and undertook testing against the three major rust diseases: stem rust (Puccinia graminis f. sp. tritici), stripe rust (Puccinia striiformis f. sp. tritici), and leaf rust (Puccinia triticina). In greenhouse screening, the method identified 19 NLRs effective against stem rust, some with resistance to isolate TTKSK, the highly virulent Ug99 lineage. This result doubled the number of known NLR resistance to stem rust from all prior efforts. The method also identified high numbers of NLRs effective against stripe rust and leaf rust. Lines have been further validated in field tests established by collaborator Dr. Brian Stefenson.
With NLRseekTM validated as a scaled method for identifying effective resistance in the field, 2Blades is now extending the platform to create arrays for other crops, such as legumes.